How we build personalized brain maps
Key Highlights:
- The human cortex varies in shape, size, and physical distortion between individuals.
- Tracing white matter fibers enables refinement of the mapping of the cerebral cortex.
- Using this method, the Structural Connectivity Atlas (SCA) allows for detailed and personalized brain maps to be generated for each individual, enabling new possibilities in precision medicine.
The Structural Connectivity Atlas (SCA)
Brain mapping has generally been a one-size-fits-all approach, however, we live in a world of 8 billion unique brains.
Understanding the connections of an individual's brain has long been shrouded in mystery. Now, for the first time, accessing precise patient-specific brain maps, regardless of brain shape, size, or physical distortion is possible. Using data about an individual's white matter connections, the SCA helps us map the location of their grey matter.
More details in the video and article below:
Introduction
Much like how the nations of Earth can be mapped on an atlas, so too can functional regions of the human brain. Compared to cartographers, however, brain mappers face a unique challenge: there isn’t one object like Earth to map, rather billions of individual brains each unified by a common set of functional regions, but separated by subtle differences in size, shape, and brain activity. Some may have also encountered previous trauma that drastically changed their brain’s structure such as brain tumor or stroke.
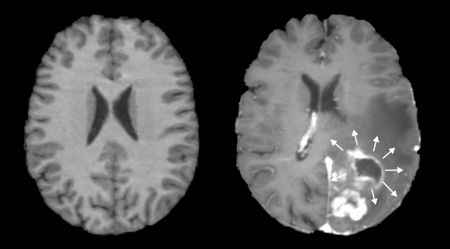
Figure 1. A normal (left), and anatomically distorted (right) human brain. Note on the right that tumor presence shifts adjacent tissue with its mass effect.
For researchers, a single generalized “brain atlas” is incredibly valuable to study the human brain at large, but this is much harder to apply at an individual level - a key limiting step to enable personalized and precision therapy for suffering patients with neurological illnesses.
Omniscient's "Stuctural Connectivity Atlas (SCA)"
Omniscient’s Structural Connectivity Atlas (SCA) is a newly developed brain mapping technique, which uses machine learning and tractographic techniques to create highly specific and personalized maps of the human brain.1
Based on an atlas developed by the Human Connectome Project (HCP)2, the SCA comprises 360 cortical areas, or parcels and adds 19 subcortical components3 to provide complete coverage of the human brain and brainstem in 379 unique parcels. This is then combined with a key refinement algorithm which identifies the location of parcels based on the specific structural characteristics of an individual’s brain. Because of this, the SCA is a substantial milestone in increasing the clinical utility and relevance of brain mapping.
Put simply, the SCA makes connectomic analysis practical at an individual level.
How the SCA works
While major white matter tract bundles within the human cortex are largely homogenous in terms of their location and shape, the location of human gray matter areas responsible for coordinating tasks and functions can differ greatly based on the size, shape, and uniformity of an individual's brain.4,5
The core principle of the SCA is that while no two brains are alike, there are common patterns in the way white matter tracts connect parcels together. This means that each parcel should have a predictable “connectivity signature” that can be used to identify it.
The SCA was constructed using three steps: training on healthy brain data, extracting white matter data from patient brain data, and applying the patterns learned on healthy data to the patient data to refine the location of gray matter parcels.
Training requires a large cohort of healthy brain data, without any structural damage or malformation. When applied to healthy brains, the locations and density of white matter tracts, which originate and terminate in gray matter parcels are studied by the SCA to determine the layout of each gray matter region in the atlas.
By first generating a map through Constrained Spherical Deconvolution (CSD), the SCA assesses the location and density of where these tracts originate and terminate - producing a “Structural Connectivity Vector”. The endpoints of white matter tracts are found in small volumes, or “voxels”, of functional gray matter parcels. By assessing these qualities of the white matter, the SCA algorithm generates a pattern for identifying each individual parcel in the atlas, based on the white matter which join them.
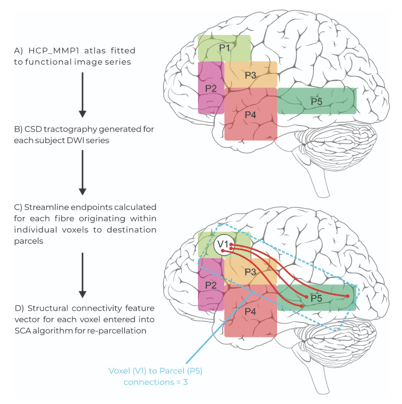
Figure 2: The extracting and application steps of the Structural Connectivity Atlas (SCA) Algorithm. Patient tractography data is entered into the model and voxels containing white matter endpoints are assigned to parcels within the modified human connectome project atlas based on their structural connectivity vector through re-parcellation.
With this pattern identified, the extracting step requires patient brain data. Once again CSD is conducted, and tractography is generated representing the endpoints and density of all white matter tracts within the brain. With patient tractography generated, the SCA asks, “which parcel is this voxel most likely to belong to, given the number of tracts which terminate and originate here?”. An important point of difference to the training step is that rather than collecting information and learning a pattern to identify gray matter, the application step appoints gray matter voxels as their most likely parcel, and in doing so “refines” the atlas from healthy brain data onto patient data in its native space.
Additionally, due to being constrained to a starting set of 379 parcels, the SCA generates reproducible results which can be discussed between patients. In sum, despite the specific locations of gray matter parcels being refined based on individual patient tractography, the end result is a personalized version of the same 379 parcel atlas, enabling valid comparisons for further analysis.
Introduction
Omniscient's "Structural connectivity Atlas (SCA)"
How the SCA works
Applying the SCA
Applying the SCA
Importantly, the SCA was built with a focus on real neurological care, and thereby incorporates features often seen with neurosurgical planning. Most notably, in cases of invasive tumours or surgical resection which has left patients without specific cortical parcels, the SCA is able to identify this empty space, and maps the remaining gray matter surrounding this tumor into its most likely atlas region. This is conceptually bound to the fact that the SCA uses white matter to map out the gray matter. That is, when structural damage of the brain leads to gray matter being destroyed, the white matter endpoints that would typically be connected to those gray matter voxels are also unable to be traced to completion - and so the SCA leaves these voxels without a classified parcel.
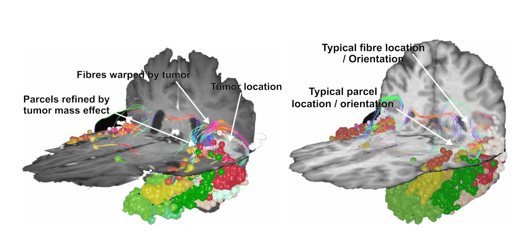
Figure 3. Gray matter re-parcellation through the SCA algorithm. By assigning gray matter voxels to parcels based on the white matter tracts which connect them, the SCA is able to effectively navigate spaces shifted or left empty by the presence or resection of a tumor.
Distinct from other atlas techniques which would require manual tuning to remove tumor-affected space, or which simply classify over a tumor site, re-parcellation allows the SCA to dynamically map an individual’s brain accounting for structural damage in the schema and nomenclature utilized in both clinical and research settings.
The SCA is only one of the techniques Omniscient delivers for performing subject-specific brain analytics. Interested in the foundational principles of modern atlassing? Check out our articles on Constrained Spherical Deconvolution and the History of Brain Mapping to learn more, or extend your knowledge further by learning more about the seven major brain networks classified within the SCA.
*Read more about the SCA in our recently published article in Human Brain Mapping
Reference list
- Doyen, S., Nicholas, P., Poologaindran, A., Crawford, L., Young, I. M., Romero-Garcia, R., & Sughrue, M. E. (2021). Connectivity-based parcellation of normal and anatomically distorted human cerebral cortex. Human Brain Mapping, 1–12. doi:10.1002/hbm. 25728
- Glasser, M.F., Coalson, T.S., Robinson, E.C., Hacker, C.D., Harwell, J., Yacoub, E., et al. (2016). A multi-modal parcellation of human cerebral cortex. Nature 536, 171-178.
- Fischl, B. (2012). FreeSurfer. Neuroimage 62, 774-781.
-
Baker, C. M., Burks, J. D., Briggs, R. G., Conner, A. K., Glenn, C. A., Sali, G., et al. (2018). A Connectomic Atlas of the Human Cerebrum-Chapter 1: Introduction, Methods, and Significance. Oper Neurosurg (Hagerstown) 15, S1-s9. doi:10.1093/ons/opy253.
-
Yeo, B. T. T., Krienen, F. M., Sepulcre, J., Sabuncu, M. R., Lashkari, D., Hollinshead, M., et al. (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology 106, 1125. doi:10.1152/JN.00338.2011.


